

(Mouseover the matches for more details )
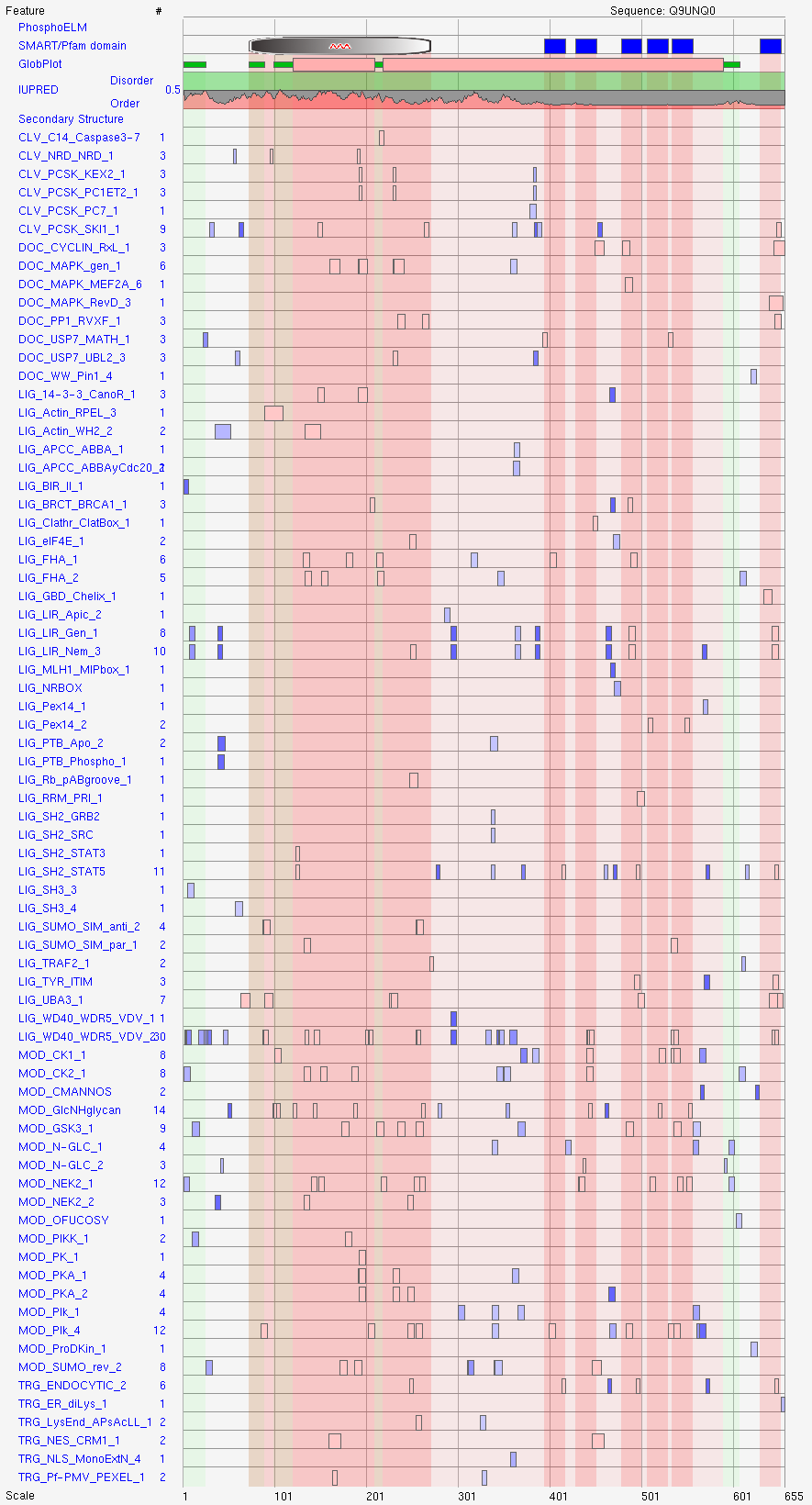
 Homologous sequences were used to calculate a multiple sequence alignment visualizing the conservation of individual short linear motifs.
Homologous sequences were used to calculate a multiple sequence alignment visualizing the conservation of individual short linear motifs.
No user supplied cellular location.
User supplied taxon: root
(An ELM is listed as filtered when all its matching instances have been filtered out.)
|
|
| Domain | Start | End |
|---|---|---|
| AAA | 72 | 270 |
| transmembrane_domain | 394 | 416 |
| transmembrane_domain | 428 | 450 |
| transmembrane_domain | 478 | 499 |
| transmembrane_domain | 506 | 528 |
| transmembrane_domain | 533 | 555 |
| transmembrane_domain | 629 | 651 |
| Elm Name | Instances (Matched Sequence) |
Positions | View in Jmol | Elm Description | Cell Compartment | Pattern | PHI-Blast Instance Mapping | Structural Filter Info | Probability | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLV_NRD_NRD_1 |
|
|
|
N-Arg dibasic convertase (NRD/Nardilysin) cleavage site (X-|-R-K or R-|-R-X). | extracellular, Golgi apparatus, cell surface |
(.RK)|(RR[^KR]) | - | - | 7.465e-03 | ||||||||||||||||||||||||||||||||||||||||||
| CLV_PCSK_KEX2_1 |
|
|
|
Yeast kexin 2 cleavage site (K-R-|-X or R-R-|-X). | extracellular, Golgi apparatus |
[KR]R. | - | - | 7.973e-03 | ||||||||||||||||||||||||||||||||||||||||||
| CLV_PCSK_PC1ET2_1 |
|
|
|
NEC1/NEC2 cleavage site (K-R-|-X). | extracellular, Golgi apparatus, Golgi membrane |
KR. | - | - | 3.903e-03 | ||||||||||||||||||||||||||||||||||||||||||
| CLV_PCSK_PC7_1 |
|
|
|
Proprotein convertase 7 (PC7, PCSK7) cleavage site (R-X-X-X-[RK]-R-|-X). | extracellular, Golgi apparatus, Golgi membrane |
R...[KR]R. | - | - | 5.087e-04 | ||||||||||||||||||||||||||||||||||||||||||
| CLV_PCSK_SKI1_1 |
|
|
|
Subtilisin/kexin isozyme-1 (SKI1) cleavage site ([RK]-X-[hydrophobic]-[LTKF]-|-X). | endoplasmic reticulum lumen, endoplasmic reticulum, Golgi apparatus, extracellular |
[RK].[AILMFV][LTKF]. | - | - | 6.821e-03 | ||||||||||||||||||||||||||||||||||||||||||
| DOC_MAPK_gen_1 |
|
|
|
MAPK interacting molecules (e.g. MAPKKs, substrates, phosphatases) carry docking motif that help to regulate specific interaction in the MAPK cascade. The classic motif approximates (R/K)xxxx#x# where # is a hydrophobic residue. | nucleus, cytosol |
[KR]{0,2}[KR].{0,2}[KR].{2,4}[ILVM].[ILVF] | - | - | 4.324e-03 | ||||||||||||||||||||||||||||||||||||||||||
| DOC_USP7_MATH_1 |
|
|
|
The USP7 MATH domain binding motif variant based on the MDM2 and p53 interactions. | nucleus | [PA][^P][^FYWIL]S[^P] | - | - | 1.239e-02 | ||||||||||||||||||||||||||||||||||||||||||
| DOC_USP7_UBL2_3 |
|
|
|
The USP7 CTD domain binding motif variant based on the ICP0 and DNMT1 interactions | nucleus | K...K | - | - | 3.742e-03 | ||||||||||||||||||||||||||||||||||||||||||
| DOC_WW_Pin1_4 |
|
|
|
The Class IV WW domain interaction motif is recognised primarily by the Pin1 phosphorylation-dependent prolyl isomerase. | cytosol, nucleus |
...([ST])P. | - | - | 1.543e-02 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_14-3-3_CanoR_1 |
|
|
|
Canonical Arg-containing phospho-motif mediating a strong interaction with 14-3-3 proteins. | nucleus, internal side of plasma membrane, cytosol |
R[^DE]{0,2}[^DEPG]([ST])(([FWYLMV].)|([^PRIKGN]P)|([^PRIKGN].{2,4}[VILMFWYP])) | - | - | 4.477e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_Actin_WH2_2 |
|
|
|
The WH2 motif is of variable length (16-19 amino acids) binding to the hydrophobic cleft formed by actin's subdomains 1 and 3. At the N-terminus it forms an alpha-helix followed by a flexible loop stabilised upon actin binding. | cytosol | [^R]..((.[ILMVF])|([ILMVF].))[^P][^P][ILVM].{4,7}L(([KR].)|(NK))[VATIGS] | - | - | 6.603e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_APCC_ABBA_1 |
|
|
|
Amphipathic motif that is involved in APC/C inhibition by binding of CDH1/CDC20. In metazoan cyclin A, the motif also acts as a degron, enabling the cyclin's degradation in prometaphase. | spindle pole, nucleus, cytosol |
[ILVMF].[ILMVP][FHY].[DE] | - | - | 3.843e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_APCC_ABBAyCdc20_2 |
|
|
|
Amphipathic motif that binds to yeast Cdc20 and acts as an APC/C degron enabling cyclin Clb5 degradation during mitosis. | not annotated | [KR]..[ILVM][FHY].[DE] | - | - | 1.669e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_BIR_II_1 |
|
|
|
These IBMs are found in pro-apoptotic proteins and function in the abrogation of caspase inhibition by Inhibitor of Apoptosis Proteins (IAPs) in apoptotic cells. The motif binds specifically to type II BIR domains. | cytosol, mitochondrion |
^M{0,1}[AS]... | - | - | 3.252e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_BRCT_BRCA1_1 |
|
|
|
Phosphopeptide motif which directly interacts with the BRCT (carboxy-terminal) domain of the Breast Cancer Gene BRCA1 with low affinity | nucleus, BRCA1-BARD1 complex |
.(S)..F | - | - | 1.912e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_eIF4E_1 |
|
|
|
Motif binding to the dorsal surface of eIF4E. | cytosol | Y....L[VILMF] | - | - | 1.891e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_FHA_1 |
|
|
|
Phosphothreonine motif binding a subset of FHA domains that show a preference for a large aliphatic amino acid at the pT+3 position. | nucleus | ..(T)..[ILV]. | - | - | 8.662e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_FHA_2 |
|
|
|
Phosphothreonine motif binding a subset of FHA domains that have a preference for an acidic amino acid at the pT+3 position. | nucleus, Replication fork |
..(T)..[DE]. | - | - | 8.286e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_LIR_Apic_2 |
|
|
|
Apicomplexa specific variant of the canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | cytosol, cytoplasmic side of late endosome membrane |
[EDST].{0,2}[WFY]..P | - | - | 3.371e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_LIR_Gen_1 |
|
|
|
Canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | cytosol, cytoplasmic side of late endosome membrane |
[EDST].{0,2}[WFY]..[ILV] | - | - | 5.200e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_LIR_Nem_3 |
|
|
|
Nematode-specific variant of the canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | cytosol, cytoplasmic side of late endosome membrane |
[EDST].{0,2}[WFY]..[ILVFY] | - | - | 6.362e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_MLH1_MIPbox_1 |
|
|
|
Proteins involved in DNA repair and replication employ conserved MIP-box motifs to bind the C-terminal domain of mismatch repair protein MLH1. | MutLalpha complex, nucleus |
.S.[FY][F] | - | - | 6.212e-05 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_NRBOX |
|
|
|
The nuclear receptor box motif (LXXLL) confers binding to nuclear receptors. | nucleus | [^P]L[^P][^P]LL[^P] | - | - | 2.628e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_Pex14_1 |
|
|
|
Wxxx[FY] motifs present in N-terminal half of Pex5 bind to Pex13 and Pex14 at peroxisomal and glycosomal membranes to facilitate entrance of PTS1 cargo proteins into the organellar lumen. | peroxisome, cytosol, glycosome |
W...[FY] | - | - | 2.226e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_PTB_Apo_2 |
|
|
|
These phosphorylation-independent motifs bind to Dab-like PTB domains. Binding is not driven by contacts at the 0 or FY position, but instead is dependent upon the large number of hydrophobic and hydrogen bond contacts between motif and domain. | integrin, internal side of plasma membrane, cytosol, receptor complex, cytoplasmic membrane-bounded vesicle |
(.[^P].NP.[FY].)|(.[ILVMFY].N..[FY].) | - | - | 3.108e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_PTB_Phospho_1 |
|
|
|
This phosphorylation-dependent motif binds to Shc-like and IRS-like PTB domains. The pTyr is positioned within a highly basic-charged anchoring pocket. A hydrophobic residue -5 (compared to pY) increases the affinity of the interaction. | integrin, internal side of plasma membrane, cytosol, receptor complex, cytoplasmic membrane-bounded vesicle |
(.[^P].NP.(Y))|(.[ILVMFY].N..(Y)) | - | - | 1.352e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_SH2_GRB2 |
|
|
|
GRB2-like Src Homology 2 (SH2) domains binding motif. | Early endosome, cytosol |
(Y)([EDST]|[MLIVAFYQ])N. | - | - | 3.019e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_SH2_SRC |
|
|
|
Src-family Src Homology 2 (SH2) domains binding motif. | cytosol | (Y)[QDEVAIL][DENPYHI][IPVGAHS] | - | - | 8.729e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_SH2_STAT5 |
|
|
|
STAT5 Src Homology 2 (SH2) domain binding motif. | cytosol | (Y)[VLTFIC].. | - | - | 3.296e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_SH3_3 |
|
|
|
This is the motif recognized by those SH3 domains with a non-canonical class I recognition specificity | plasma membrane, focal adhesion, cytosol |
...[PV]..P | - | - | 1.317e-02 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_SH3_4 |
|
|
|
This is the motif recognized by those SH3 domains with a non-canonical class II recognition specificity | focal adhesion, cytosol |
KP..[QK]... | - | - | 6.781e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_TRAF2_1 |
|
|
|
Major TRAF2-binding consensus motif. Members of the tumor necrosis factor receptor (TNFR) superfamily initiate intracellular signaling by recruiting the C-domain of the TNFR-associated factors (TRAFs) through their cytoplasmic tails. | cytosol | [PSAT].[QE]E | - | - | 4.300e-03 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_TYR_ITIM |
|
|
|
ITIM (immunoreceptor tyrosine-based inhibitory motif). Phosphorylation of the ITIM motif, found in the cytoplasmic tail of some inhibitory receptors (KIRs) that bind MHC Class I, leads to the recruitment and activation of a protein tyrosine phosphatase. | cytosol | [ILV].(Y)..[ILV] | - | - | 2.992e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_WD40_WDR5_VDV_1 |
|
|
|
This WDR5-binding motif binds to a cleft between blades 5 and 6 of the WD40 repeat domain of WDR5, opposite of the Win motif-binding site, to mediate assembly of histone modification complexes. | nucleus, histone methyltransferase complex |
[ED].{0,3}[VIL]D[VI] | - | - | 2.939e-04 | ||||||||||||||||||||||||||||||||||||||||||
| LIG_WD40_WDR5_VDV_2 |
|
|
|
Fungi-specific variant of the WDR5-binding motif that binds to a cleft between blades 5 and 6 of the WD40 repeat domain of WDR5, opposite of the Win motif-binding site, to mediate assembly of histone modification complexes. | nucleus, histone methyltransferase complex |
[EDSTY].{0,4}[VIPLA][TSDEKR][ILVA] | - | - | 4.678e-02 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_CK1_1 |
|
|
|
CK1 phosphorylation site | cytosol, nucleus |
S..([ST])... | - | - | 1.704e-02 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_CK2_1 |
|
|
|
CK2 phosphorylation site | nucleus, cytosol, protein kinase CK2 complex |
...([ST])..E | - | - | 1.457e-02 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_CMANNOS |
|
|
|
Motif for attachment of a mannosyl residue to a tryptophan | extracellular | (W)..W | - | - | 4.692e-05 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_GlcNHglycan |
|
|
|
Glycosaminoglycan attachment site | extracellular, Golgi apparatus |
[ED]{0,3}.(S)[GA]. | - | - | 1.792e-02 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_GSK3_1 |
|
|
|
GSK3 phosphorylation recognition site | cytosol, nucleus |
...([ST])...[ST] | - | - | 2.679e-02 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_N-GLC_1 |
|
|
|
Generic motif for N-glycosylation. It was shown that Trp, Asp, and Glu are uncommon before the Ser/Thr position. Efficient glycosylation usually occurs when ~60 residues or more separate the glycosylation acceptor site from the C-terminus. | extracellular, Golgi apparatus, endoplasmic reticulum |
.(N)[^P][ST].. | - | - | 5.018e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_N-GLC_2 |
|
|
|
Atipical motif for N-glycosylation site. Examples are Human CD69, which is uniquely glycosylated at typical (Asn-X-Ser/Thr) and atypical (Asn-X-Cys) motifs, beta protein C | extracellular, Golgi apparatus, endoplasmic reticulum |
(N)[^P]C | - | - | 2.973e-04 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_NEK2_1 |
|
|
|
NEK2 phosphorylation motif with preferred Phe, Leu or Met in the -3 position to compensate for less favorable residues in the +1 and +2 position. | centrosome, Ndc80 complex, condensed nuclear chromosome outer kinetochore, cytosol, nucleus |
[FLM][^P][^P]([ST])[^DEP][^DE] | - | - | 9.798e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_NEK2_2 |
|
|
|
NEK2 phosphorylation motif with specific set of residues in the +1 and +2 position to compensate for less favorable residues in the -3 position. | centrosome, Ndc80 complex, condensed nuclear chromosome outer kinetochore, cytosol, nucleus |
[WYPCAG][^P][^P]([ST])[IFCVML][KRHYF] | - | - | 1.295e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_OFUCOSY |
|
|
|
Site for attachment of a fucose residue to a serine. | extracellular | C.{3,5}([ST])C | - | - | 4.617e-05 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_PIKK_1 |
|
|
|
(ST)Q motif which is phosphorylated by PIKK family members. | nucleus, cytosol |
...([ST])Q.. | - | - | 9.230e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_PKA_1 |
|
|
|
Main preference for PKA-type AGC kinase phosphorylation. | cAMP-dependent protein kinase complex, cytosol, nucleus |
[RK][RK].([ST])[^P].. | - | - | 2.315e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_PKA_2 |
|
|
|
Secondary preference for PKA-type AGC kinase phosphorylation. | cytosol, nucleus, cAMP-dependent protein kinase complex |
.R.([ST])[^P].. | - | - | 9.458e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_Plk_1 |
|
|
|
Ser/Thr residue phosphorylated by the Plk1 kinase | centralspindlin complex, nucleus, spindle, gamma-tubulin complex, midbody, cytosol, kinetochore, spindle midzone, nuclear condensin complex, cleavage furrow, nucleoplasm, microtubule organizing center |
.[DNE][^PG][ST](([FYILMVW]..)|([^PEDGKN][FWYLIVM]).) | - | - | 7.674e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_Plk_4 |
|
|
|
Ser/Thr residue phosphorylated by Plk4 | nucleus, cytosol, SCF ubiquitin ligase complex, cleavage furrow, centriole, gamma-tubulin ring complex, centriolar satellite, pericentriolar material |
..[^IRFW]([ST])[ILMVFWY][ILMVFWY]. | - | - | 6.019e-03 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_ProDKin_1 |
|
|
|
Proline-Directed Kinase (e.g. MAPK) phosphorylation site in higher eukaryotes. | cytosol, nucleus |
...([ST])P.. | - | - | 1.543e-02 | ||||||||||||||||||||||||||||||||||||||||||
| MOD_SUMO_rev_2 |
|
|
|
Inverted version of SUMOylation motif recognized for modification by SUMO-1 | PML body, nucleus |
[SDE].{0,5}[DE].(K).{0,1}[AIFLMPSTV] | - | - | 1.280e-02 | ||||||||||||||||||||||||||||||||||||||||||
| TRG_ENDOCYTIC_2 |
|
|
|
Tyrosine-based sorting signal responsible for the interaction with mu subunit of AP (Adaptor Protein) complex | plasma membrane, clathrin-coated endocytic vesicle, cytosol |
Y..[LMVIF] | - | - | 2.587e-03 | ||||||||||||||||||||||||||||||||||||||||||
| TRG_ER_diLys_1 |
|
|
|
ER retention and retrieving signal found at the C-terminus of type I ER membrane proteins (cytoplasmic in this topology). Di-Lysine signal is responsible for COPI-mediated retrieval from post-ER compartments. | endoplasmic reticulum membrane, integral protein, ER-Golgi transport vesicle membrane, endoplasmic reticulum membrane, Golgi-ER transport vesicle membrane, rough endoplasmic reticulum, endoplasmic reticulum, endoplasmic reticulum cisterna, COPI coated vesicle membrane, cytosol |
K.{0,1}K.{2,3}$ | - | - | 2.677e-05 | ||||||||||||||||||||||||||||||||||||||||||
| TRG_LysEnd_APsAcLL_1 |
|
|
|
Sorting and internalisation signal found in the cytoplasmic juxta-membrane region of type I transmembrane proteins. Targets them from the Trans Golgi Network to the lysosomal-endosomal-melanosomal compartments. Interacts with adaptor protein (AP) complexes | cytosol, Endocytic vesicle |
[DERQ]...L[LVI] | - | - | 2.758e-03 | ||||||||||||||||||||||||||||||||||||||||||
| TRG_NLS_MonoExtN_4 |
|
|
|
Monopartite variant of the classical basically charged NLS. N-extended version. | nucleus, Nuclear pore, NLS-dependent protein nuclear import complex |
(([PKR].{0,1}[^DE])|([PKR]))((K[RK])|(RK))(([^DE][KR])|([KR][^DE]))[^DE] | - | - | 1.276e-03 | ||||||||||||||||||||||||||||||||||||||||||
| TRG_Pf-PMV_PEXEL_1 |
|
|
|
Plasmodium Export Element, PEXEL, is a trafficking signal for protein cleavage by PMV protease and export from Plasmodium parasites to infected host cells. | endoplasmic reticulum, host cell cytoplasm, extracellular, host cell outer membrane, pathogen-containing vacuole membrane |
(R.[LI].[EDQ])|(R.L..[EDQ])|(K.L.E) | - | - | 2.161e-03 |
| Elm Name | Positions | View in Jmol | Elm Description | Cell Compartment | Pattern | PHI-Blast Instance Mapping | Structural Filter Info | Probability | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLV_C14_Caspase3-7 |
|
|
Caspase-3 and Caspase-7 cleavage site. | cytosol, nucleus |
[DSTE][^P][^DEWHFYC]D[GSAN] | - | - | 3.094e-03 | ||||||||||||||||||||||||||||||||
| CLV_NRD_NRD_1 |
|
|
N-Arg dibasic convertase (NRD/Nardilysin) cleavage site (X-|-R-K or R-|-R-X). | extracellular, Golgi apparatus, cell surface |
(.RK)|(RR[^KR]) | - | - | 7.465e-03 | ||||||||||||||||||||||||||||||||
| CLV_PCSK_KEX2_1 |
|
|
Yeast kexin 2 cleavage site (K-R-|-X or R-R-|-X). | extracellular, Golgi apparatus |
[KR]R. | - | - | 7.973e-03 | ||||||||||||||||||||||||||||||||
| CLV_PCSK_PC1ET2_1 |
|
|
NEC1/NEC2 cleavage site (K-R-|-X). | extracellular, Golgi apparatus, Golgi membrane |
KR. | - | - | 3.903e-03 | ||||||||||||||||||||||||||||||||
| CLV_PCSK_SKI1_1 |
|
|
Subtilisin/kexin isozyme-1 (SKI1) cleavage site ([RK]-X-[hydrophobic]-[LTKF]-|-X). | endoplasmic reticulum lumen, endoplasmic reticulum, Golgi apparatus, extracellular |
[RK].[AILMFV][LTKF]. | - | - | 6.821e-03 | ||||||||||||||||||||||||||||||||
| DOC_CYCLIN_RxL_1 |
|
|
This motif is mainly based on cyclin A binding peptides and may not apply to all cyclins. | nucleus, membrane, cytosol, centriole, Cajal body |
(.|([KRH].{0,3}))[^EDWNSG][^D][RK][^D]L.{0,1}[FLMP].{0,3}[EDST] | - | - | 4.211e-03 | ||||||||||||||||||||||||||||||||
| DOC_MAPK_gen_1 |
|
|
MAPK interacting molecules (e.g. MAPKKs, substrates, phosphatases) carry docking motif that help to regulate specific interaction in the MAPK cascade. The classic motif approximates (R/K)xxxx#x# where # is a hydrophobic residue. | nucleus, cytosol |
[KR]{0,2}[KR].{0,2}[KR].{2,4}[ILVM].[ILVF] | - | - | 4.324e-03 | ||||||||||||||||||||||||||||||||
| DOC_MAPK_MEF2A_6 |
|
|
A kinase docking motif that mediates interaction towards the ERK1/2 and p38 subfamilies of MAP kinases. | nucleus, cytosol, Transcription factor complex |
[RK].{2,4}[LIVMP].[LIV].[LIVMF] | - | - | 2.584e-03 | ||||||||||||||||||||||||||||||||
| DOC_MAPK_RevD_3 |
|
|
Reverse (C to N direction) of the classical MAPK docking motif ELM:DOC_MAPK_gen_1 with an often extended linker region of the bipartite motif. | cytosol, nucleus |
[LIVMPFA].[LIV].{1,2}[LIVMP].{4,6}[LIV]..[RK][RK] | - | - | 1.805e-04 | ||||||||||||||||||||||||||||||||
| DOC_PP1_RVXF_1 |
|
|
Protein phosphatase 1 catalytic subunit (PP1c) interacting motif binds targeting proteins that dock to the substrate for dephosphorylation. The motif defined is [RK]{0,1}[VI][^P][FW]. | nucleus, protein phosphatase type 1 complex, cytosol |
..[RK].{0,1}[VIL][^P][FW]. | - | - | 8.301e-04 | ||||||||||||||||||||||||||||||||
| DOC_USP7_MATH_1 |
|
|
The USP7 MATH domain binding motif variant based on the MDM2 and p53 interactions. | nucleus | [PA][^P][^FYWIL]S[^P] | - | - | 1.239e-02 | ||||||||||||||||||||||||||||||||
| DOC_USP7_UBL2_3 |
|
|
The USP7 CTD domain binding motif variant based on the ICP0 and DNMT1 interactions | nucleus | K...K | - | - | 3.742e-03 | ||||||||||||||||||||||||||||||||
| LIG_14-3-3_CanoR_1 |
|
|
Canonical Arg-containing phospho-motif mediating a strong interaction with 14-3-3 proteins. | nucleus, internal side of plasma membrane, cytosol |
R[^DE]{0,2}[^DEPG]([ST])(([FWYLMV].)|([^PRIKGN]P)|([^PRIKGN].{2,4}[VILMFWYP])) | - | - | 4.477e-03 | ||||||||||||||||||||||||||||||||
| LIG_Actin_RPEL_3 |
|
|
RPEL motif, present in proteins in several repeats, mediates binding to the hydrophobic cleft created by subdomains 1 and 3 of G-actin. | cytosol | [IL]..[^P][^P][^P][^P]R.....[IL]..[^P][^P][ILV][ILM] | - | - | 6.116e-06 | ||||||||||||||||||||||||||||||||
| LIG_Actin_WH2_2 |
|
|
The WH2 motif is of variable length (16-19 amino acids) binding to the hydrophobic cleft formed by actin's subdomains 1 and 3. At the N-terminus it forms an alpha-helix followed by a flexible loop stabilised upon actin binding. | cytosol | [^R]..((.[ILMVF])|([ILMVF].))[^P][^P][ILVM].{4,7}L(([KR].)|(NK))[VATIGS] | - | - | 6.603e-04 | ||||||||||||||||||||||||||||||||
| LIG_BRCT_BRCA1_1 |
|
|
Phosphopeptide motif which directly interacts with the BRCT (carboxy-terminal) domain of the Breast Cancer Gene BRCA1 with low affinity | nucleus, BRCA1-BARD1 complex |
.(S)..F | - | - | 1.912e-03 | ||||||||||||||||||||||||||||||||
| LIG_Clathr_ClatBox_1 |
|
|
Clathrin box motif found on cargo adaptor proteins, it interacts with the beta propeller structure located at the N-terminus of Clathrin heavy chain. | cytosol, Golgi apparatus, cytoskeleton, clathrin-coated endocytic vesicle, Golgi trans-face |
L[IVLMF].[IVLMF][DE] | - | - | 3.406e-04 | ||||||||||||||||||||||||||||||||
| LIG_eIF4E_1 |
|
|
Motif binding to the dorsal surface of eIF4E. | cytosol | Y....L[VILMF] | - | - | 1.891e-04 | ||||||||||||||||||||||||||||||||
| LIG_FHA_1 |
|
|
Phosphothreonine motif binding a subset of FHA domains that show a preference for a large aliphatic amino acid at the pT+3 position. | nucleus | ..(T)..[ILV]. | - | - | 8.662e-03 | ||||||||||||||||||||||||||||||||
| LIG_FHA_2 |
|
|
Phosphothreonine motif binding a subset of FHA domains that have a preference for an acidic amino acid at the pT+3 position. | nucleus, Replication fork |
..(T)..[DE]. | - | - | 8.286e-03 | ||||||||||||||||||||||||||||||||
| LIG_GBD_Chelix_1 |
|
|
Amphipatic alpha helix that binds the GTPase-binding domain (GBD) in WASP and N-WASP. | extracellular exosome, vesicle membrane, cytosol, actin cytoskeleton, cell-cell junction |
[ILV][VA][^P][^P][LI][^P][^P][^P][LM] | - | - | 9.792e-05 | ||||||||||||||||||||||||||||||||
| LIG_LIR_Gen_1 |
|
|
Canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | cytosol, cytoplasmic side of late endosome membrane |
[EDST].{0,2}[WFY]..[ILV] | - | - | 5.200e-03 | ||||||||||||||||||||||||||||||||
| LIG_LIR_Nem_3 |
|
|
Nematode-specific variant of the canonical LIR motif that binds to Atg8 protein family members to mediate processes involved in autophagy. | cytosol, cytoplasmic side of late endosome membrane |
[EDST].{0,2}[WFY]..[ILVFY] | - | - | 6.362e-03 | ||||||||||||||||||||||||||||||||
| LIG_Pex14_2 |
|
|
Fxxx[WF] motifs are present in Pex19 and S. cerevisiae Pex5 cytosolic receptors that bind to peroxisomal membrane docking member, Pex14 | cytosol, peroxisome, glycosome |
F...[WF] | - | - | 4.628e-04 | ||||||||||||||||||||||||||||||||
| LIG_Rb_pABgroove_1 |
|
|
The LxxLFD motif binds in a deep groove between pocket A and pocket B of the Retinoblastoma protein | nucleus, Transcription factor complex, Rb-E2F complex |
..[LIMV]..[LM][FY]D. | - | - | 2.475e-05 | ||||||||||||||||||||||||||||||||
| LIG_RRM_PRI_1 |
|
|
The PTB RRM2 Interacting (PRI) motif is found in some splicing regulators, possibly only in the chordate lineage. As part of splicing complex regulation, it interacts with the 2nd RNA binding domain (RRM) of PTB, the polypyrimidine tract binding protein. | nucleus, heterogeneous nuclear ribonucleoprotein complex |
.[ILVM]LG..P. | - | - | 8.391e-05 | ||||||||||||||||||||||||||||||||
| LIG_SH2_STAT3 |
|
|
YXXQ motif found in the cytoplasmic region of cytokine receptors that bind STAT3 SH2 domain. | cytosol | (Y)..Q | - | - | 7.975e-04 | ||||||||||||||||||||||||||||||||
| LIG_SH2_STAT5 |
|
|
STAT5 Src Homology 2 (SH2) domain binding motif. | cytosol | (Y)[VLTFIC].. | - | - | 3.296e-03 | ||||||||||||||||||||||||||||||||
| LIG_SUMO_SIM_anti_2 |
|
|
Motif for the antiparallel beta augmentation mode of non-covalent binding to SUMO protein. | PML body, nucleus, nuclear body |
[DEST]{1,10}.{0,1}[VIL][DESTVILMA][VIL][VILM].[DEST]{0,5} | - | - | 2.349e-03 | ||||||||||||||||||||||||||||||||
| LIG_SUMO_SIM_par_1 |
|
|
Motif for the parallel beta augmentation mode of non-covalent binding to SUMO protein. | PML body, nucleus, nuclear body |
[DEST]{0,5}.[VILPTM][VIL][DESTVILMA][VIL].{0,1}[DEST]{1,10} | - | - | 4.545e-03 | ||||||||||||||||||||||||||||||||
| LIG_TRAF2_1 |
|
|
Major TRAF2-binding consensus motif. Members of the tumor necrosis factor receptor (TNFR) superfamily initiate intracellular signaling by recruiting the C-domain of the TNFR-associated factors (TRAFs) through their cytoplasmic tails. | cytosol | [PSAT].[QE]E | - | - | 4.300e-03 | ||||||||||||||||||||||||||||||||
| LIG_TYR_ITIM |
|
|
ITIM (immunoreceptor tyrosine-based inhibitory motif). Phosphorylation of the ITIM motif, found in the cytoplasmic tail of some inhibitory receptors (KIRs) that bind MHC Class I, leads to the recruitment and activation of a protein tyrosine phosphatase. | cytosol | [ILV].(Y)..[ILV] | - | - | 2.992e-04 | ||||||||||||||||||||||||||||||||
| LIG_UBA3_1 |
|
|
UBA3 adenylation domain binding motif variant based on the UBE2M and UBE2F interactions. | nucleus | [ILM][ILMF].{1,2}[ILM].{0,4}K | - | - | 1.196e-03 | ||||||||||||||||||||||||||||||||
| LIG_WD40_WDR5_VDV_2 |
|
|
Fungi-specific variant of the WDR5-binding motif that binds to a cleft between blades 5 and 6 of the WD40 repeat domain of WDR5, opposite of the Win motif-binding site, to mediate assembly of histone modification complexes. | nucleus, histone methyltransferase complex |
[EDSTY].{0,4}[VIPLA][TSDEKR][ILVA] | - | - | 4.678e-02 | ||||||||||||||||||||||||||||||||
| MOD_CK1_1 |
|
|
CK1 phosphorylation site | cytosol, nucleus |
S..([ST])... | - | - | 1.704e-02 | ||||||||||||||||||||||||||||||||
| MOD_CK2_1 |
|
|
CK2 phosphorylation site | nucleus, cytosol, protein kinase CK2 complex |
...([ST])..E | - | - | 1.457e-02 | ||||||||||||||||||||||||||||||||
| MOD_GlcNHglycan |
|
|
Glycosaminoglycan attachment site | extracellular, Golgi apparatus |
[ED]{0,3}.(S)[GA]. | - | - | 1.792e-02 | ||||||||||||||||||||||||||||||||
| MOD_GSK3_1 |
|
|
GSK3 phosphorylation recognition site | cytosol, nucleus |
...([ST])...[ST] | - | - | 2.679e-02 | ||||||||||||||||||||||||||||||||
| MOD_N-GLC_2 |
|
|
Atipical motif for N-glycosylation site. Examples are Human CD69, which is uniquely glycosylated at typical (Asn-X-Ser/Thr) and atypical (Asn-X-Cys) motifs, beta protein C | extracellular, Golgi apparatus, endoplasmic reticulum |
(N)[^P]C | - | - | 2.973e-04 | ||||||||||||||||||||||||||||||||
| MOD_NEK2_1 |
|
|
NEK2 phosphorylation motif with preferred Phe, Leu or Met in the -3 position to compensate for less favorable residues in the +1 and +2 position. | centrosome, Ndc80 complex, condensed nuclear chromosome outer kinetochore, cytosol, nucleus |
[FLM][^P][^P]([ST])[^DEP][^DE] | - | - | 9.798e-03 | ||||||||||||||||||||||||||||||||
| MOD_NEK2_2 |
|
|
NEK2 phosphorylation motif with specific set of residues in the +1 and +2 position to compensate for less favorable residues in the -3 position. | centrosome, Ndc80 complex, condensed nuclear chromosome outer kinetochore, cytosol, nucleus |
[WYPCAG][^P][^P]([ST])[IFCVML][KRHYF] | - | - | 1.295e-03 | ||||||||||||||||||||||||||||||||
| MOD_PIKK_1 |
|
|
(ST)Q motif which is phosphorylated by PIKK family members. | nucleus, cytosol |
...([ST])Q.. | - | - | 9.230e-03 | ||||||||||||||||||||||||||||||||
| MOD_PK_1 |
|
|
Phosphorylase kinase phosphorylation site | cytosol | [RK]..(S)[VI].. | - | - | 9.418e-04 | ||||||||||||||||||||||||||||||||
| MOD_PKA_1 |
|
|
Main preference for PKA-type AGC kinase phosphorylation. | cAMP-dependent protein kinase complex, cytosol, nucleus |
[RK][RK].([ST])[^P].. | - | - | 2.315e-03 | ||||||||||||||||||||||||||||||||
| MOD_PKA_2 |
|
|
Secondary preference for PKA-type AGC kinase phosphorylation. | cytosol, nucleus, cAMP-dependent protein kinase complex |
.R.([ST])[^P].. | - | - | 9.458e-03 | ||||||||||||||||||||||||||||||||
| MOD_Plk_4 |
|
|
Ser/Thr residue phosphorylated by Plk4 | nucleus, cytosol, SCF ubiquitin ligase complex, cleavage furrow, centriole, gamma-tubulin ring complex, centriolar satellite, pericentriolar material |
..[^IRFW]([ST])[ILMVFWY][ILMVFWY]. | - | - | 6.019e-03 | ||||||||||||||||||||||||||||||||
| MOD_SUMO_rev_2 |
|
|
Inverted version of SUMOylation motif recognized for modification by SUMO-1 | PML body, nucleus |
[SDE].{0,5}[DE].(K).{0,1}[AIFLMPSTV] | - | - | 1.280e-02 | ||||||||||||||||||||||||||||||||
| TRG_ENDOCYTIC_2 |
|
|
Tyrosine-based sorting signal responsible for the interaction with mu subunit of AP (Adaptor Protein) complex | plasma membrane, clathrin-coated endocytic vesicle, cytosol |
Y..[LMVIF] | - | - | 2.587e-03 | ||||||||||||||||||||||||||||||||
| TRG_LysEnd_APsAcLL_1 |
|
|
Sorting and internalisation signal found in the cytoplasmic juxta-membrane region of type I transmembrane proteins. Targets them from the Trans Golgi Network to the lysosomal-endosomal-melanosomal compartments. Interacts with adaptor protein (AP) complexes | cytosol, Endocytic vesicle |
[DERQ]...L[LVI] | - | - | 2.758e-03 | ||||||||||||||||||||||||||||||||
| TRG_NES_CRM1_1 |
|
|
Some proteins re-exported from the nucleus contain a Leucine-rich nuclear export signal (NES) binding to the CRM1 exportin protein. | nucleus, cytosol |
([DEQ].{0,1}[LIM].{2,3}[LIVMF][^P]{2,3}[LMVF].[LMIV].{0,3}[DE])|([DE].{0,1}[LIM].{2,3}[LIVMF][^P]{2,3}[LMVF].[LMIV].{0,3}[DEQ]) | - | - | 7.626e-04 | ||||||||||||||||||||||||||||||||
| TRG_Pf-PMV_PEXEL_1 |
|
|
Plasmodium Export Element, PEXEL, is a trafficking signal for protein cleavage by PMV protease and export from Plasmodium parasites to infected host cells. | endoplasmic reticulum, host cell cytoplasm, extracellular, host cell outer membrane, pathogen-containing vacuole membrane |
(R.[LI].[EDQ])|(R.L..[EDQ])|(K.L.E) | - | - | 2.161e-03 |